ImageJ: Filters#
Introduction#
Most of the filters we’ve considered are available through the submenu. This section adds a little more information about their implementation in ImageJ, and asks a few questions.
Linear filters#
Mean filters#
The easiest way to apply a 3×3 mean filter in ImageJ is through the command. The fact that the shortcut is Shift+S can almost make this too easy, as I find myself accidentally smoothing when I really wanted to save my image. Take care.
To apply larger mean filters, the command is . It uses approximately circular neighborhoods, and the neighborhood size is adjusted by choosing a Radius value. The command displays the neighborhoods used for different values of Radius. If you happen to choose Radius = 1, you get a 3×3 filter – and the same results as using .

Gaussian filters#
is the command that implements a Gaussian filter.
In the event that you want a Gaussian filter that isn’t isotropic (i.e. has a different size along different dimensions), can be used.
Although not really recommended, unsharp masking is available through .
Difference of Gaussians
There’s currently no direct command in ImageJ to implement difference of Gaussians filtering, rather the steps need to be pieced together with image duplication and subtraction. However Difference of Gaussians describes how to generate a macro for DoG filtering.
Custom linear filters#
makes it possible to define any custom linear filter by entering the values of the desired coefficients, separated by spaces and arranged in rows and columns. If you Normalize Kernel is selected, then the coefficients are scaled so that they add to 1, by dividing by the sum of all the coefficients – unless the sum is 0, in which case requesting normalizion does nothing.
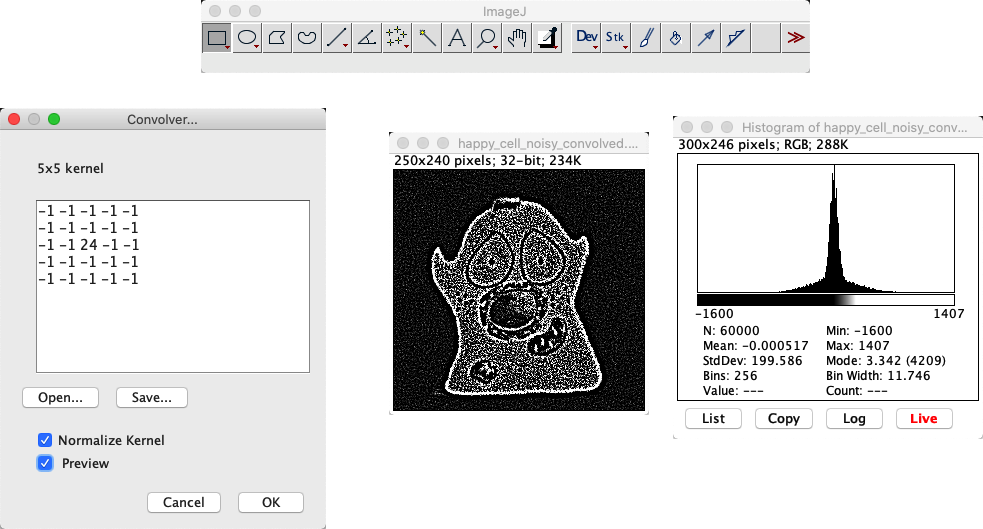
When defining an n_×_n filter kernel with , ImageJ insists that n is an odd number. Why?
If n is an odd number, the filter has a clear central pixel. This makes it possible to center the filter kernel on a pixel on the image.
Predict what happens when you convolve an image using a filter that consists of a single coefficient with a value -1 in the following cases:
Normalize Kernel is checked
You have a 32-bit image, Normalize Kernel is unchecked
You have an 8-bit image, Normalize Kernel is unchecked
The results of convolving with a single -1 coefficient in different circumstances:
Normalize Kernel is checked: Nothing at all happens. The normalization makes the filter just a single 1… and convolving with a single 1 leaves the image unchanged.
You have a 32-bit image (Normalize Kernel unchecked): The pixel values become negative, and the image looks inverted.
You have an 8-bit image (Normalize Kernel unchecked): The pixel values would become negative, but then cannot be stored in an 8-bit unsigned integer form. Therefore, all pixels simply become clipped to zero.
Using any image, work out which of the methods for dealing with boundaries shown in Fig. 86 is used by ImageJ’s command.
Note: This requires a bit of creativity. It will certainly help to use an image with some variation at the image boundary. I used .
Replication of boundary pixels is the default method used by in ImageJ (although other filtering plugins by different authors might use different methods).
My approach to test this involved using with a filter that consisting of a 1 followed by a lot of zeros (e.g. 1 0 0 0 0 0 0 0 0 0 0 0 0...).
This basically shifts the image to the right, bringing whatever is outside the image boundary into view.
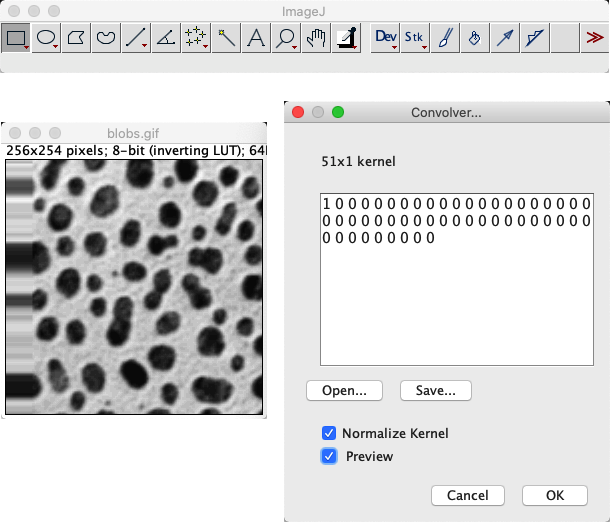

Fig. 100 Gradient magnitude#
Practice using the commands we’ve met so far by determining the gradient magnitude of an image, as described here.
You will need to use
Several commands in the submenu
Something else we’ve used before… possibly
If you need a sample image, you can use . (Be sure to pay attention to the bit-depth!)
The process to calculate the gradient magnitude is:
Convert the image to 32-bit (if it isn’t already 32-bit)
Duplicate the image
Convolve one copy of the image with the horizontal gradient filter, and one with the vertical (i.e. coefficients
-1 0 1arranged as a row or column)Compute the square of both images ()
Use the image calculator to add the images together
Compute the square root of the resulting image ()
Here’s a macro that implements these steps:
run("32-bit");
id1 = getImageID()
run("Duplicate...", " ");
id2 = getImageID();
run("Convolve...", "text1=[-1 0 1\n] normalize");
run("Square");
selectImage(id1);
run("Convolve...", "text1=-1\n0\n1\n normalize");
run("Square");
imageCalculator("Add create", id1, id2);
run("Square Root");
The convolution results in negative values, which is why the 32-bit conversion is needed.
Note: This is (almost) what is done by the command , except the gradient filters are slightly different.

Fig. 101 The ‘Edges’ LUT#
ImageJ has a LUT called edges under . Applied to , it does a rather good job of highlighting edges – without actually changing the pixels at all.
How does it work? Does it apply a filter?
The LUT shows most low and high pixel values as black – and uses lighter shades of gray only for a small range of values in between (see ). In any image with a good separation of background and foreground pixels, but which still has a somewhat smooth transition between them, this means everything but the edges can appear black.
All this is achieved by a LUT: no pixels were harmed, there was no filtering applied.
Nonlinear filters#
Rank filters#
The main rank filters are to be found exactly where you might expect them:
ImageJ uses circular neighborhoods with its built-in rank filters, similar to how mean filters are implemented. We will meet these filters again in Morphological operations.
Removing outliers#
Fig. 88 shows that median filtering is much better than mean filtering for removing outliers. We might encounter this if something in the microscope is not quite functioning as expected or if dark noise is a problem, but otherwise we expect the noise in fluorescence microscopy images to produce few really extreme values (see Noise).
Nevertheless, provides an alternative if isolated bright values are present. This is a nonlinear filter that inserts median values only whenever a pixel is found that is further away from the local median than some adjustable threshold.
It’s therefore like a more selective median filter that will only modify the image at pixels where it is considered really necessary.

